Cytokines sit at the crossroads of almost every immune-driven process—from early inflammatory signaling to late-stage tissue remodeling. For CROs supporting discovery or translational studies, cytokine readouts have shifted from being "supportive biomarkers" to primary endpoints that define study outcomes.
In practical terms, this means assay selection is no longer a technical afterthought. The platform you choose determines how well your data capture the biological complexity of immune responses. A mismatch between study design and detection method can lead to wasted samples, inconsistent QC results, or even ambiguous conclusions.
Modern multiplex technologies—Luminex, MSD, and Simoa—each bring distinct analytical philosophies:
- Luminex prioritizes breadth, allowing simultaneous measurement of dozens of cytokines with minimal sample input.
- MSD (Meso Scale Discovery) balances sensitivity with matrix tolerance, offering electrochemiluminescent precision suitable for complex biological fluids.
- Simoa (Single Molecule Array) delivers single-molecule-level detection for targets that sit at the edge of detectability.
For a CRO team, the decision among these systems extends beyond instrument performance. It touches study logistics, cost structure, and downstream interpretability. The goal is to ensure every dataset—whether exploratory or confirmatory—rests on a fit-for-purpose analytical foundation.
This article translates those platform capabilities into a practical decision framework, showing how project design, biological expectations, and analytical trade-offs align to produce reliable cytokine data.
Luminex vs MSD vs Simoa: Technical Overview
| Feature | Luminex xMAP | MSD (ECL) | Simoa (Digital ELISA) |
|---|---|---|---|
| Detection Principle | Fluorescent bead multiplexing | Electrochemiluminescence | Digital counting of single complexes |
| Multiplex Capacity | Up to 50 targets | 10–20 targets | 3–10 targets |
| Sensitivity | pg/mL | sub-pg to mid-pg | fg/mL (ultra-sensitive) |
| Matrix Tolerance | Moderate | High | Moderate with careful blocking |
| Best Use Case | Large-scale profiling | Quantitative bridging studies | Low-abundance validation |
Luminex suits exploratory work where breadth and efficiency matter most. MSD provides balanced precision for regulated or cross-matrix studies. Simoa excels when targets are rare or sample volume is limited.
Each platform answers a different research question—the art lies in choosing the one that aligns with your project's purpose. For CROs, understanding these trade-offs ensures that platform selection supports both scientific accuracy and operational efficiency.
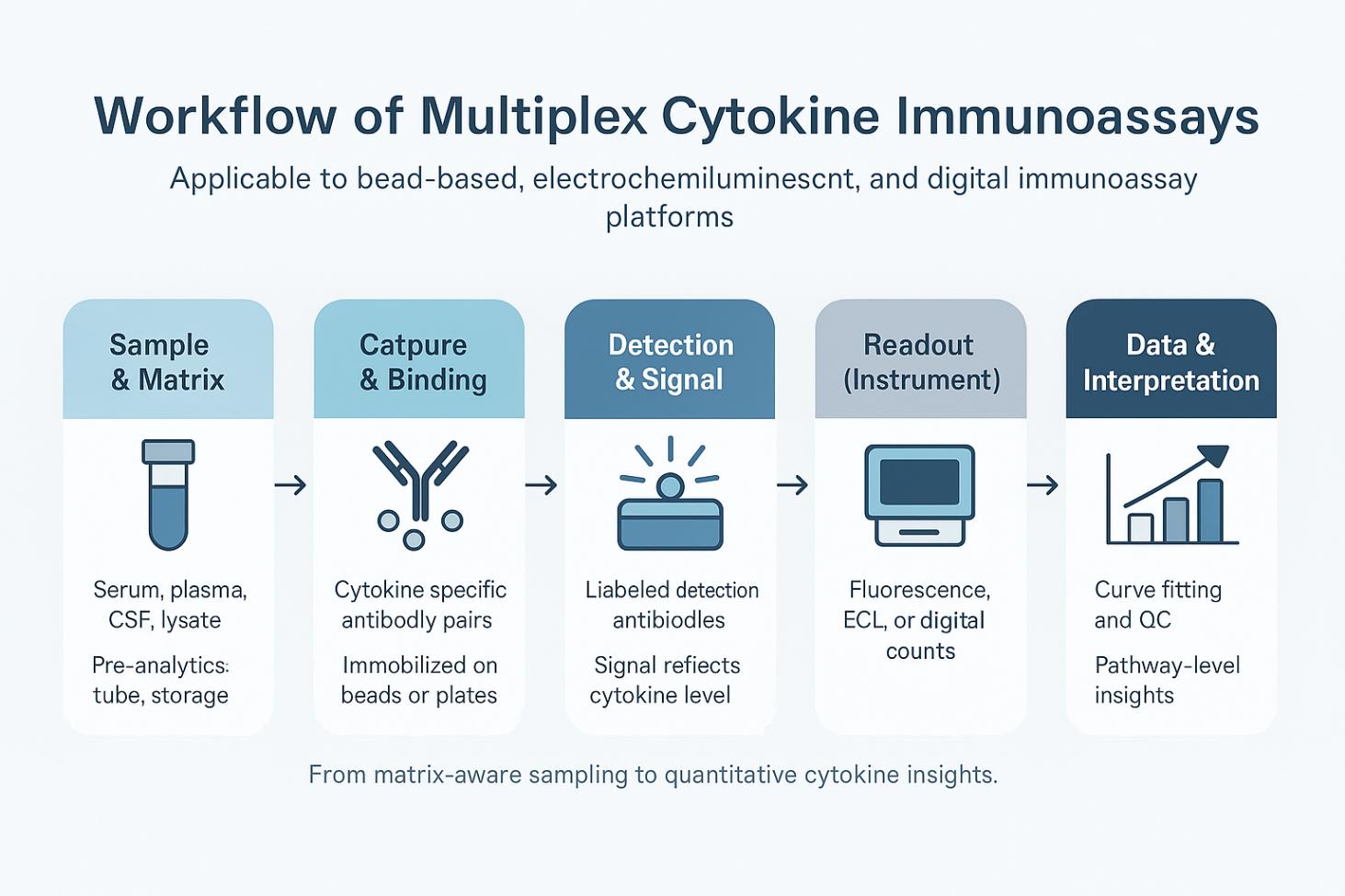 Workflow of multiplex cytokine immunoassays.
Workflow of multiplex cytokine immunoassays.
Choosing the Right Platform: From Study Design to Practical Trade-offs
Selecting a cytokine detection platform is rarely a question of brand—it's a question of fit. Every study sits somewhere along a spectrum: from exploratory discovery to confirmatory validation. The right platform depends on where your project lies on that curve, how much biological signal you expect, and how constrained your sample volume or budget might be.
Match the Platform to the Scientific Scenario
Each technology serves a distinct analytical purpose:
- Luminex – For High-Throughput Exploration
When broad screening is required across dozens of cytokines, Luminex offers the flexibility and cost-efficiency to handle large cohorts. It's ideal for early immunoprofiling, toxicology panels, or large comparative screens where throughput and multiplexing are the main priorities. - MSD – For Balanced Quantification Across Complex Matrices
MSD's electrochemiluminescence (ECL) detection bridges sensitivity and robustness. It performs consistently in serum, CSF, or tissue lysates and maintains linearity across several logs. It is often chosen for translational projects or bridging studies that require reproducibility and traceable QC records. - Simoa – For Precision in Low-Abundance Targets
When cytokines approach the limits of conventional assays, Simoa's single-molecule digital readout delivers the necessary sensitivity. It's particularly useful for rare analytes, early-stage disease models, or studies using very limited samples.
Each of these systems excels when its analytical strengths align with the project's biological question.
Balance Analytical Depth With Practical Realities
Beyond technical fit, the decision often reflects the study's priorities—how deep to measure versus how broad to explore.
| Project Focus | Recommended Platform Approach | Primary Strengths |
|---|---|---|
| Exploratory Discovery | Luminex or MSD | Large panel coverage, low sample volume, cost-efficient data scaling |
| Targeted Validation | MSD | Balanced sensitivity, robust performance in complex matrices |
| Precision or Confirmatory Studies | Simoa | Ultra-sensitive quantification, ideal for low-abundance cytokines |
| Integrated Design | Luminex / MSD → Simoa | Two-phase strategy: screen broadly, then confirm with high sensitivity |
This framework helps teams make objective trade-offs: it's not about choosing the best platform, but the most appropriate one for that point in the research pipeline.
Think in Phases, Not Silos
In modern immunology workflows, many CROs use a hybrid design—leveraging different technologies at different stages. A typical pattern is to screen with a broad Luminex or MSD panel to capture system-wide changes, then use Simoa for confirmatory analysis of key low-abundance markers. This approach maximizes data depth while keeping the study design lean and cost-rational.
Practical Guidance for Platform Selection
The most effective cytokine programs begin by aligning assay capability with biological intent. Sensitivity or throughput alone rarely define success—how those factors serve the study's core question does.
When evaluating Luminex, MSD, or Simoa, consider three anchors:
- the biological mechanism you aim to characterize,
- the sample and resource limits you must work within,
- and the degree of reproducibility your data must support.
By framing the choice through these lenses, the right platform often becomes self-evident and defensible.
RFP & SOW Essentials: How to Define Your Multiplex Cytokine Study Clearly
Many cytokine projects run into challenges not because of assay performance, but because the initial study scope wasn't precisely described. When requests for proposals or work statements leave room for interpretation, small technical gaps can turn into large data inconsistencies later.
Below are practical points that help research sponsors and CRO teams communicate clearly from the beginning—ensuring the analytical setup truly reflects the study's intent.
Clarify the Panel Objectives
Start by defining what the cytokine data should reveal.
Is the goal to explore immune activation broadly, or to verify a focused set of biomarkers?
Listing the specific analytes and their biological context helps both parties align on realistic expectations. Even minor differences—such as IL-12p70 versus total IL-12—can affect platform suitability and cost structure.
Tip: Specify whether flexibility in analyte substitution is acceptable, especially if one platform doesn't support all intended targets.
Describe the Biological Context, Not Just the Matrix
Instead of simply stating "serum" or "plasma," include short notes about how samples were obtained and stored.
Details such as collection tube type, anticoagulant, or storage duration may influence assay reproducibility.
These notes guide the CRO in selecting compatible panels or recommending proper validation steps—without adding any extra burden on your end.
Align on Detection Expectations
A single phrase like "high sensitivity" can mean different things across technologies.
Whenever possible, mention the expected concentration range for key cytokines or the biological model you're working with.
This helps narrow down the platform choice:
- Broader multiplex screening → typically Luminex or MSD
- Ultra-low detection targets → likely Simoa or equivalent sensitivity
The goal is to match assay dynamic range to biological signal, not to brand preferences.
Outline Quality Benchmarks Up Front
Different labs define "acceptable QC" differently.
Agreeing on general quality principles avoids misunderstandings later.
You might include expectations such as:
- Clear standard curves and replicates within an acceptable variation range
- Basic control samples or dilution checks
- Transparent documentation of outlier handling
This doesn't have to be a rigid checklist—think of it as a shared quality language between sponsor and service team.
Discuss Batching and Consistency
Cytokine measurements are sensitive to subtle changes in environment and reagent lots. If samples are processed over multiple runs, it's helpful to decide early whether to use reference controls or calibration bridging between batches. Such consistency planning improves the interpretability of results, especially when studies extend over longer periods or involve multiple collaborators.
Define Data Deliverables for Reuse
When the project ends, the most valuable product isn't just the results table—it's the ability to reuse and verify that data later. Request clear, organized files such as:
- Raw signals and curve-fit parameters
- Plate maps and QC summaries
- Final normalized concentration tables
Structured deliverables make it easier to integrate cytokine results with transcriptomics, proteomics, or pharmacodynamic datasets down the road.
Keep Communication Transparent
Regular progress sharing and review checkpoints are often underestimated. Agreeing on update frequency and format—such as periodic data reviews or summary dashboards—helps maintain alignment without constant back-and-forth. Clear communication also accelerates problem-solving when analytical or logistical questions arise.
Ensuring Data Quality and Cross-Platform Reproducibility
When cytokine results drive go/no-go decisions, "good enough" isn't enough. Below is the quality architecture we recommend and routinely implement so your findings remain defensible across plates, batches, and platforms (Luminex, MSD, Simoa).
A Three-Layer QC Architecture
Plate-level controls (intra-plate)
- Blank, ≥7-point standard curve (4PL/5PL), technical duplicates, HQC/LQC.
- Layout avoids edge effects; high-range markers include planned dilution lanes.
Within-batch controls
- Pooled matrix QC prepared from study-matched samples; placed on every plate to monitor drift and precision.
Across-batch / cross-site controls
- Bridging controls (a fixed mini-panel of samples or references) rerun across batches/sites to anchor long-term comparability.
What this gives you: transparent, plate-to-project traceability and an auditable trail for every reported value.
 Three-layer QC architecture for multiplex cytokine studies.
Three-layer QC architecture for multiplex cytokine studies.
Curve Fitting, Reportable Range, and Outlier Logic
- Model: 4PL or 5PL; asymmetry favors 5PL. Weighting (1/y or 1/y²) to reduce high-end leverage.
- Back-calculation: typical acceptance around ±20% RE (lowest standard may allow a slightly wider band); any refit is versioned and explained.
- Reportable range: concentrations are only reported within LLOQ–ULOQ per plate; <LLOQ is not "forced quantified."
- Precision: replicate %CV typically targeted at ≤15–20%, with low-end flexibility and explicit flags.
- Outliers: robust rules (e.g., MAD) mark and document influence before/after exclusion.
What this gives you: concentration values that can be reconstructed from raw signal, with defensible acceptance logic.
Matrix Effects, Parallelism, and Recovery
- Parallelism: diluted sample slope within ~0.9–1.1 of the calibrator is considered acceptable; divergence triggers buffer/ blocking optimization or platform reconsideration.
- Spike-recovery: target ~80–120%; directional bias prompts reagent or matrix strategy review.
- Pre-analytical transparency: anticoagulant, hemolysis/lipemia flags, freeze–thaw counts are recorded with the data to support covariate modeling.
What this gives you: confidence that platform readouts reflect biology, not matrix artifacts.
Handling <LLOQ, >ULOQ, and Missingness
- <LLOQ: carried as "<LLOQ"; statistical exports include optional half-LLOQ or ROS imputation columns for sensitivity analyses.
- >ULOQ: re-assayed after dilution when available; otherwise flagged as ">ULOQ" and kept separate from quantified values.
- Missingness classes: technical failure vs. insufficient volume vs. protocol exclusion—tracked distinctly to detect bias.
What this gives you: clean downstream analysis without hidden imputation decisions.
Plate Normalization and Batch Effect Control
- QC-anchored drift correction: pooled-QC signals anchor per-plate scaling; temporal drift modeled (e.g., linear/LOESS) and versioned.
- Statistical harmonization: for large cohorts or multi-omics joins, options include ComBat or mixed-effects modeling with batch as a random factor.
- Before/after deliverables: Levey–Jennings and density plots show the effect of any correction.
What this gives you: reproducible data that travel well—across time, teams, and analyses.
Cross-Platform Bridging (Luminex ↔ MSD ↔ Simoa)
- Sampling: ≥30 representative samples spanning low/mid/high.
- Method comparison: Deming or Passing–Bablok regression, plus Bland–Altman for bias and range dependence.
- Alignment: constant bias—apply single/multi-point factors; non-linear bias—segment or re-anchor LLOQ.
- Orthogonal checks: for sensitive markers (e.g., IFN-α, IL-17 family), confirm directionality on a second platform.
What this gives you: continuity of evidence even when technologies differ.
What You Receive (Data Package)
- Raw signals (MFI/ECL/digital counts), curve parameters, curve images.
- Plate maps, reagent lot IDs, instrument/software versions.
- QC summary (HQC/LQC, pooled QC, positive/negative controls, parallelism, recovery).
- Versioned analysis notes and "before/after" correction files.
- Change log for any reruns, exclusions, or refits.
What this gives you: a complete evidence chain—from signal to decision—ready for internal review or external audit.
How Creative Proteomics Helps
Creative Proteomics provides end-to-end multiplex cytokine analysis using the Luminex xMAP platform as our core technology, complemented by MSD and Simoa systems when higher sensitivity or specific matrix performance is needed.
Our scientists help clients design panels that match their targets, validate assay performance through multi-layer QC, and deliver complete, transparent data packages for downstream use.
By integrating throughput, precision, and sensitivity across platforms, we offer fit-for-purpose cytokine profiling that supports confident decisions in immunology, oncology, and translational research.
FAQ
What's the key difference between Luminex, MSD, and Simoa for multiplex cytokines?
Luminex is bead-based multiplexing that reads color-coded microspheres to quantify many cytokines efficiently in one small sample; MSD uses electrochemiluminescence (ECL) on patterned electrodes to achieve low background and broad linear range in complex matrices; Simoa is a digital ELISA that counts single immunocomplexes in femtoliter wells, pushing sensitivity into the fg/mL domain for very low-abundance targets.
How is multiplex immunoassay different from traditional single-plex ELISA?
Multiplex immunoassays measure many analytes simultaneously to maximize information per sample, but they demand stronger method checks—such as spike-recovery and dilution linearity—to confirm that matrix effects or cross-reactivity aren't biasing any member of the panel, whereas single-plex ELISA validates one target at a time.
What is "parallelism" and why does it matter in cytokine assays?
Parallelism tests whether a diluted sample's response curve runs in parallel with the calibrator curve; when slopes/shape align, the assay is quantifying proportionally across dilutions (matrix interference is controlled), and when they diverge, teams adjust diluent or blocking—or reconsider platform/antibody pairing—to restore proportionality.
What is the hook (prozone) effect and how do teams mitigate it?
At very high antigen (or antibody) levels, sandwich immunoassays can under-report due to disrupted complex formation—this "hook" effect is mitigated by planned high-dilution channels and curve-shape monitoring, because extreme concentrations can yield falsely low or negative results across platforms if not checked.
Do I need Simoa's ultra-sensitivity for every cytokine panel?
No—Simoa is most valuable when expected concentrations sit near or below typical pg/mL ranges (e.g., rare cytokines or early signals), whereas high-plex discovery or matrix-tolerant quantitation often favors Luminex or MSD respectively; many programs screen broadly first, then reserve Simoa for confirmatory low-end targets.
Which platforms are better for challenging matrices like serum, CSF, or tissue lysates?
ECL on MSD plates is widely used for complex matrices because repeated electrical excitation and low optical background support wide dynamic range and stable quantitation, while bead-based Luminex provides high multiplexing for profiling and Simoa is chosen when ultra-low signals must be resolved within those matrices.
Can data from Luminex, MSD, and Simoa be combined in one study?
Yes—method-comparison workflows typically include sample coverage across low/mid/high ranges and use Deming or Passing–Bablok regression plus Bland–Altman analysis to characterize bias and agreement before applying any alignment factors, with documentation preserved for auditability.
What should I look for in spike-and-recovery results when selecting a platform or kit?
Acceptable spike-and-recovery indicates the matrix and reagents are compatible (the spiked analyte is measured close to its known value), whereas directional bias suggests matrix interference or capture/detection chemistry issues that should be tuned via diluent, blocking, or kit selection prior to large-scale runs.
When is multiplex preferable to running many single-plex assays?
Multiplex is preferable when sample volume is scarce or when pathway-level interpretation needs concurrent measurement of many cytokines, because one-tube, many-marker designs increase information density and reduce handling variability—provided validation (parallelism, recovery, cross-reactivity control) is in place.
How do platform physics influence decision-making beyond "sensitivity vs. throughput"?
Bead optics (Luminex), ECL kinetics on electrodes (MSD), and digital counting at the single-molecule level (Simoa) each shape precision, matrix tolerance, and low-end detectability; mapping these physics to your expected concentration range and matrix often clarifies which system will generate the most defensible cytokine evidence.
References:
- Vignali, D. A. A. "Multiplexed particle-based flow cytometric assays." Journal of Immunological Methods 243.1–2 (2000): 243–255.
- Rissin, J. L., et al. "Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations." Nature Biotechnology 28.6 (2010): 595–599.
- Chowdhury, F., Williams, A., Johnson, P. "Validation and comparison of two multiplex technologies, Luminex® and Mesoscale Discovery®, for human cytokine profiling." Journal of Immunological Methods 340.1 (2009): 55–64.
- Bastarache, J. A., Koyama, T., Wickersham, N. E., et al. "Validation of a multiplex electrochemiluminescent immunoassay platform in human and mouse samples." Journal of Immunological Methods 408 (2014): 13–23.
- Lee, J. W., Devanarayan, V., Barrett, Y. C., et al. "Fit-for-Purpose Method Development and Validation for Successful Biomarker Measurement." Pharmaceutical Research 23.2 (2006): 312–328.