Cytokines are small, soluble proteins that act as critical mediators of cell signaling, primarily within the immune system. They regulate numerous biological processes, including inflammation, immune response modulation, cell proliferation, differentiation, and apoptosis. Given their central role in immune regulation, cytokines have emerged as key targets in the development of therapeutics for autoimmune diseases, cancer, and inflammatory disorders.
In the early stages of drug development, particularly in basic research and preclinical studies, understanding cytokine signaling pathways provides essential insights into disease mechanisms and therapeutic opportunities. Advances in molecular biology, structural biology, and computational modeling have enabled researchers to dissect these complex pathways, identify novel drug targets, and design molecules with improved specificity and efficacy.
This article explores how cytokine signaling pathways influence drug discovery, focusing on target identification, lead compound development, and preclinical evaluation. By understanding the molecular intricacies of cytokine networks, researchers can design therapies that precisely modulate pathological immune responses while minimizing off-target effects. The following sections will provide a detailed overview of the structural and functional aspects of cytokine pathways, methods for discovering and validating therapeutic targets, and strategies for lead optimization and preclinical testing.
Cytokine Signaling Pathways for Drug Discovery
Structural and Functional Insights into Cytokines and Receptors
Cytokines exert their biological effects by binding to specific cell surface receptors, triggering downstream signaling cascades. Structurally, cytokines are categorized into several families based on their molecular architecture and receptor interactions:
- Interleukins (ILs): Essential regulators of immune cell differentiation and activation. IL-6 and IL-17, for instance, are prominent targets in autoimmune disease therapy.
- Interferons (IFNs): Critical for antiviral defense and immune surveillance. IFN-α and IFN-β are used in viral infection and cancer therapies.
- Tumor Necrosis Factors (TNFs): Mediators of inflammation and apoptosis. TNF-α inhibitors, like infliximab, are widely used in rheumatoid arthritis and Crohn's disease.
- Colony-Stimulating Factors (CSFs): Promote hematopoiesis and immune cell differentiation, often targeted in bone marrow recovery therapies.
Receptor-Ligand Interactions
Cytokine receptors are generally classified into:
- Type I and Type II Cytokine Receptors: Often associated with JAK-STAT signaling, these receptors play key roles in immune regulation.
- TNF Receptors (TNFRs): Involved in apoptosis and inflammation. Structural insights into TNFR-ligand complexes have facilitated the design of small molecule inhibitors.
- Interleukin Receptors: Complex receptor subunits that induce diverse signaling responses. Therapeutics like IL-6 receptor antibodies are prominent in autoimmune disease treatment.
Structural biology techniques, such as X-ray crystallography and cryo-electron microscopy (cryo-EM), have elucidated high-resolution cytokine-receptor complexes. This structural information is invaluable for structure-based drug design (SBDD) and the identification of small molecule inhibitors, peptide mimetics, or biologics that block cytokine-receptor interactions.
Moreover, advances in computational docking and molecular dynamics simulations allow for the in silico screening of potential drug candidates, expediting early-stage drug discovery.
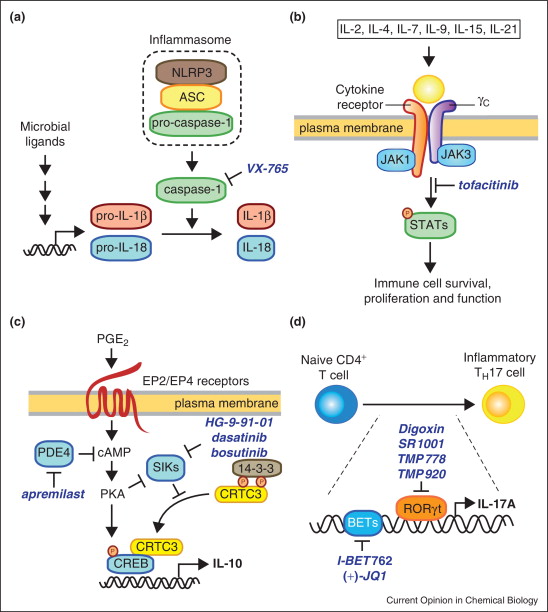 Representative pathways regulating cytokine production and signaling that have been targeted by small molecules (Sundberg, Thomas B et al., 2014).
Representative pathways regulating cytokine production and signaling that have been targeted by small molecules (Sundberg, Thomas B et al., 2014).
Intracellular Signaling Pathways
Upon cytokine binding, intracellular signaling cascades are triggered, leading to gene expression changes that regulate immune responses. Several key pathways are particularly relevant for drug development:
JAK-STAT Pathway
The Janus kinase (JAK)-Signal Transducer and Activator of Transcription (STAT) pathway is one of the most well-studied cytokine signaling mechanisms. Cytokines such as IL-6, IFN-γ, and IL-12 activate JAKs, which phosphorylate STAT proteins. The phosphorylated STATs dimerize and translocate to the nucleus to regulate gene expression.
JAK inhibitors (e.g., tofacitinib, ruxolitinib) have shown success in treating autoimmune diseases like rheumatoid arthritis and myelofibrosis. Rational design of next-generation JAK inhibitors with improved selectivity remains a major focus in preclinical research.
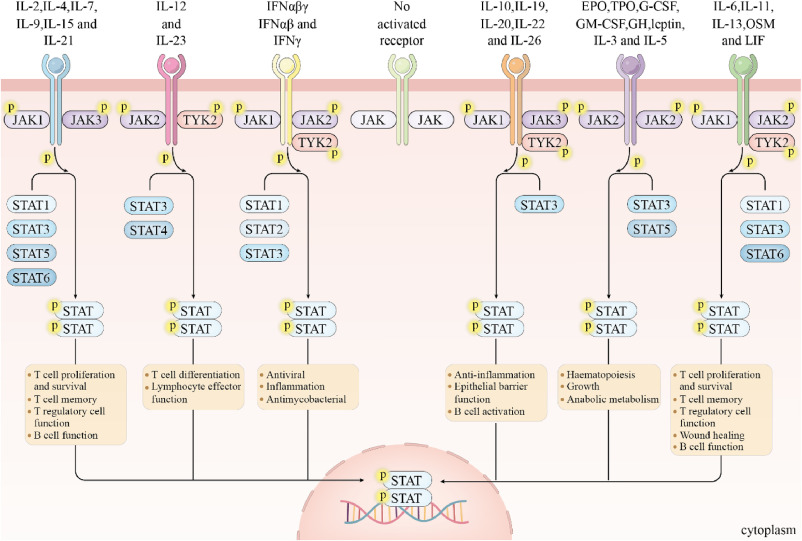 Cytokine signaling through the JAK-STAT pathway in physiological conditions (Sarapultsev et al., 2023).
Cytokine signaling through the JAK-STAT pathway in physiological conditions (Sarapultsev et al., 2023).
NF-κB Pathway
Activated by cytokines like TNF-α and IL-1, the NF-κB pathway regulates inflammatory responses. Inhibitors of IκB kinase (IKK), a key activator of NF-κB, are under investigation for conditions involving chronic inflammation.
Preclinical studies focus on selective IKK inhibitors and peptide-based NF-κB pathway blockers to reduce off-target effects. Understanding pathway cross-talk is crucial for optimizing therapeutic outcomes.
MAPK and PI3K-AKT Pathways
Mitogen-activated protein kinase (MAPK) and phosphoinositide 3-kinase (PI3K)-AKT pathways are often activated downstream of cytokine receptors, influencing cell proliferation, differentiation, and survival.
Inhibitors targeting components like MEK, ERK, or PI3K are under preclinical evaluation for autoimmune diseases and cancers. Dual-pathway inhibitors are also being explored to enhance therapeutic efficacy.
Systems Biology and Network Analysis
Cytokine signaling pathways are not isolated; they form interconnected networks with extensive cross-talk and feedback regulation. Systems biology approaches provide comprehensive insights into these complex interactions, enhancing drug discovery efforts.
Computational Modeling
Mathematical models and simulations are used to predict the dynamic behavior of cytokine networks. By integrating multi-omics data (e.g., genomics, proteomics, transcriptomics), researchers can simulate the effects of pathway perturbations caused by potential drug candidates.
Predictive modeling helps identify novel therapeutic targets, assess drug combination strategies, and minimize off-target effects.
Network Analysis
Protein-protein interaction networks map the connections between cytokines, receptors, and downstream effectors. Network analysis algorithms can prioritize key nodes for therapeutic targeting.
By identifying pathway hubs and feedback regulators, drug developers can design combination therapies that address compensatory mechanisms within cytokine networks.
Artificial Intelligence (AI) and Machine Learning
AI-powered algorithms are increasingly used to analyze vast biological datasets, identify biomarker signatures, and predict drug-target interactions. Machine learning models trained on cytokine signaling data can suggest novel drug candidates or optimize lead compound properties.
AI-driven drug discovery platforms accelerate the identification of cytokine modulators, reducing time and cost in the early stages of development.
Target Identification and Validation
Target identification and validation are critical steps in the early stages of drug development. The goal is to pinpoint specific molecules within cytokine signaling pathways that can be therapeutically modulated to alter disease progression. Cytokines and their receptors, intracellular kinases, transcription factors, and downstream effectors are all potential targets.
High-Throughput Screening (HTS) and Assay Development
High-throughput screening (HTS) is an essential method for discovering small molecule inhibitors or biologics that modulate cytokine signaling. It allows the rapid evaluation of thousands to millions of compounds in a short time, generating valuable leads for further optimization.
Assay Types for Cytokine Pathway Screening
To ensure robust and reliable results, various assay formats are developed to measure cytokine activity, receptor binding, or downstream signaling:
- Reporter Gene Assays:
Reporter constructs containing response elements from cytokine-activated transcription factors (e.g., STAT or NF-κB) linked to luciferase or fluorescent proteins are used to measure pathway activation. These assays are ideal for screening inhibitors of JAK-STAT or NF-κB pathways. - ELISA and Multiplex Cytokine Assays:
Enzyme-linked immunosorbent assays (ELISA) quantify cytokine levels in cell culture supernatants or biological fluids.
Luminex multiplex platform enables simultaneous detection of multiple cytokines, offering comprehensive pathway insights. - Cell-Based Phosphorylation Assays:
Western blotting, flow cytometry, or proximity-based assays such as AlphaLISA detect phosphorylated signaling proteins (e.g., p-STAT3, p-ERK). These assays are particularly useful for monitoring pathway inhibition. - Surface Plasmon Resonance (SPR) and Bio-Layer Interferometry (BLI):
SPR and BLI provide real-time kinetic analysis of cytokine-receptor interactions, assisting in lead compound optimization by measuring binding affinity and kinetics.
Services you may be interested in:
Target Identification Techniques
The identification of druggable targets within cytokine signaling pathways is a complex but critical process. Multiple approaches are employed to identify potential candidates:
Proteomics and Phosphoproteomics
Mass spectrometry-based proteomics quantifies cytokine-induced changes in protein abundance or post-translational modifications. Phosphoproteomics, in particular, is crucial for studying signaling cascades, identifying activated kinases, and discovering downstream effectors.
In inflammatory diseases, phosphoproteomic profiling can uncover aberrant kinase activity, leading to the identification of novel therapeutic targets.
Computational Target Discovery
Machine learning algorithms and systems biology models integrate multi-omics data to predict cytokine pathway targets. Network analysis reveals key regulators and hub proteins that are likely to be effective therapeutic targets.
Target Validation Strategies
Once a potential target is identified, rigorous validation is necessary to confirm its relevance and therapeutic potential. Various experimental approaches are applied to ensure confidence in the target selection:
Genetic Validation
- Gene Knockout and Knockdown Models:
Gene editing or RNAi-mediated knockdown can be used to assess the effect of target inhibition on cytokine signaling. Loss-of-function studies in disease-relevant cell lines or animal models are crucial for validating therapeutic relevance. - Overexpression Studies:
Conversely, overexpressing the target protein in cells can reveal its role in cytokine-driven disease mechanisms.
Pharmacological Validation
Small molecule inhibitors, monoclonal antibodies, or cytokine receptor antagonists can be used to block target activity. If target inhibition results in a desirable therapeutic effect, further drug development is warranted.
- Chemical Probes:
Selective, potent chemical probes with well-characterized pharmacokinetics are frequently used in preclinical studies to validate targets before progressing to drug candidates.
Biomarker Analysis
- Predictive Biomarkers:
Identifying biomarkers that correlate with target engagement and pathway inhibition is essential. Biomarkers can include phosphorylated proteins, cytokine levels, or gene expression signatures. - Patient-Derived Models:
Ex vivo analysis of patient samples using organoids, tumor slices, or immune cell cultures allows for biomarker identification and validation in a physiologically relevant context.
Lead Discovery and Optimization
Once a cytokine pathway target is identified and validated, the next phase involves discovering and optimizing lead compounds. Lead discovery typically focuses on identifying molecules with desirable pharmacological properties that can modulate the target's activity.
Approaches to Lead Discovery
Lead discovery involves identifying promising compounds that interact with the cytokine-related target. Several common approaches include:
High-Throughput Screening (HTS)
HTS allows the screening of large chemical libraries to identify inhibitors that modulate the cytokine pathway. It is a high-efficiency method, particularly effective for kinase inhibitors or cytokine-receptor interactions.
Structure-Based Drug Design (SBDD)
Using structural insights from X-ray crystallography or cryo-EM, SBDD allows for the design of molecules that fit precisely into the target's binding pocket, improving the potency and specificity of the lead compound.
Fragment-Based Drug Discovery (FBDD)
FBDD identifies smaller fragments that bind weakly to the target. These fragments are then optimized to create potent lead compounds. This approach is particularly useful for difficult-to-target cytokine receptor interactions.
Lead Optimization Strategies
Once initial leads are discovered, they are optimized for better potency, selectivity, and pharmacokinetic properties.
Potency and Selectivity Improvement
Through medicinal chemistry, SAR studies, and computational tools, lead compounds are refined to increase binding affinity and specificity to the target. This also includes exploring allosteric inhibitors, which bind outside the active site, offering higher selectivity and fewer off-target effects.
Drug-Like Properties
Leads undergo testing for ADME properties (absorption, distribution, metabolism, excretion) to ensure they meet drug-like criteria. Prodrugs or modifications are often designed to enhance stability and bioavailability, ensuring that the compound can reach therapeutic concentrations in vivo.
Toxicity and Off-Target Effects
Lead compounds are evaluated for safety through in vitro and in vivo toxicology studies. Additionally, selectivity profiles are tested to minimize off-target interactions that could cause adverse effects.
Pharmacokinetics and Pharmacodynamics (PK/PD)
Once lead compounds are optimized, their pharmacokinetics (ADME) and pharmacodynamics (target engagement and pathway inhibition) are evaluated. This helps assess their clinical potential, including their ability to reach target tissues and effectively modulate the cytokine pathways.
Preclinical Evaluation and Biomarker Development
In Vitro Evaluation
In vitro assays are the first step in preclinical testing, offering a controlled environment to assess a compound's activity against its target and downstream signaling pathways.
Target Engagement and Pathway Inhibition
- Cell-Based Assays:
Cytokine-responsive cell lines expressing the target receptor are commonly used to measure pathway activation or inhibition. Assays often monitor phosphorylation of key signaling proteins (e.g., STATs, NF-κB) using Western blotting, ELISA, or flow cytometry. - Reporter Gene Assays:
Luciferase-based reporter systems containing cytokine-responsive elements provide a quantifiable readout of pathway activity. - Ligand Binding Assays:
echniques such as surface plasmon resonance (SPR) and bio-layer interferometry (BLI) are used to determine binding affinity, kinetics, and selectivity of a compound.
Cytokine Release and Functional Effects
- Cytokine Profiling:
Multiplex immunoassays like Luminex or ELISA can quantify cytokine levels released by immune cells following compound treatment. Suppression of pro-inflammatory cytokines (e.g., TNF-α, IL-6) indicates pathway inhibition. - Cell Viability and Proliferation:
In oncology applications, cytokine blockade may induce tumor cell apoptosis or reduce proliferation, which can be assessed using cell viability assays such as MTT or Annexin V staining.
In Vivo Evaluation
Animal models are essential for evaluating the pharmacokinetics (PK), pharmacodynamics (PD), and efficacy of cytokine-targeted therapies in a complex biological system.
Disease Models
- Inflammatory and Autoimmune Disease Models:
Cytokine-driven diseases like rheumatoid arthritis and inflammatory bowel disease are often studied in mouse models using inflammatory stimuli such as lipopolysaccharide (LPS) or collagen-induced arthritis (CIA). - Cancer Models:
Xenograft or syngeneic tumor models help evaluate the effects of cytokine pathway inhibition on tumor growth, particularly in cancers driven by cytokine signaling.
Pharmacokinetics and Pharmacodynamics
- PK Studies:
Plasma and tissue samples are collected at various time points to assess compound concentration, clearance, and bioavailability. Techniques such as LC-MS/MS are commonly used for analysis. - PD Studies:
Biomarkers, including phosphorylated signaling proteins or downstream cytokine levels, are monitored to confirm target engagement and pathway inhibition.
Toxicology and Safety Assessment
- Acute and Chronic Toxicity Studies:
Multiple-dose toxicity studies in rodents and non-human primates assess potential toxic effects. Hematology, clinical chemistry, and histopathology evaluations are conducted to detect organ toxicity. - Cytokine Release Syndrome (CRS) Evaluation:
Since cytokine-targeted therapies may induce CRS, animal models are used to monitor cytokine levels and immune cell activation.
Biomarker Development
Biomarkers are essential for understanding drug mechanism of action, monitoring treatment response, and predicting efficacy or toxicity. In cytokine-targeted drug development, biomarkers can be classified into three main categories:
Predictive Biomarkers
- Target Expression Levels:
High expression of cytokine receptors or elevated cytokine levels in patient samples may predict responsiveness to targeted therapies. For instance, elevated IL-6 levels often correlate with successful response to IL-6 inhibitors. - Genomic and Transcriptomic Biomarkers:
Specific gene expression signatures associated with cytokine pathway activation can guide patient selection for targeted therapies.
Pharmacodynamic Biomarkers
- Pathway Inhibition Markers:
Monitoring downstream signaling events, such as reduced phosphorylation of STAT proteins, serves as a direct measure of target engagement. - Cytokine Modulation:
Reduced levels of inflammatory cytokines like TNF-α, IL-1β, or IL-17 in response to therapy can indicate pathway inhibition.
Safety Biomarkers
- Cytokine Release Monitoring:
Early detection of elevated IL-6, IL-1, or TNF-α levels can provide a warning of cytokine release syndrome (CRS), allowing timely management. - Immune Cell Profiling:
Flow cytometry and single-cell RNA sequencing are used to monitor immune cell populations for signs of excessive immune suppression or activation.
References:
- Sarapultsev, Alexey, et al. "JAK-STAT signaling in inflammation and stress-related diseases: implications for therapeutic interventions." Molecular biomedicine 4.1 (2023): 40. https://doi.org/10.1186/s43556-023-00151-1
- Sundberg, Thomas B., et al. "Small-molecule control of cytokine function: new opportunities for treating immune disorders." Current opinion in chemical biology 23 (2014): 23-30. https://doi.org/10.1016/j.cbpa.2014.08.013
- Yang, Lan, et al. "The signal pathways and treatment of cytokine storm in COVID-19." Signal transduction and targeted therapy 6.1 (2021): 255. https://doi.org/10.1038/s41392-021-00679-0

